Exploring the words we use to describe COVID-19
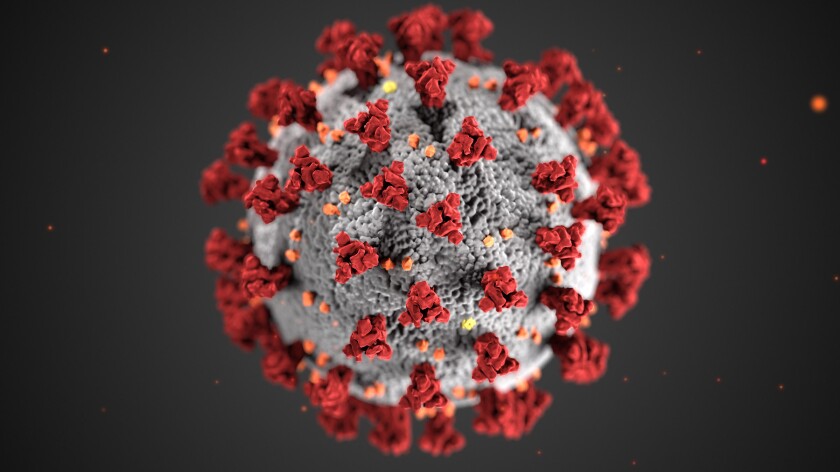
As the coronavirus clamps its tentacles around our planet and the number of infections and deaths burgeons, you might be wondering why the respiratory infection is now dubbed COVID-19.
In this instance, the method of word formation is called a clipped compound. Each component of the word is shortened and strung together. CO is a clipping of corona, VI of virus and D of disease. The 19 identifies the year the outbreak began.
Corona derives from a Greek-through-Latin word for garland, wreath or crown (as in Coronado). The name refers to the characteristic appearance, under an electron microscope, of virions, the infective form of the virus. These virions have a fringe of large, bulbous surface projections that create an image resembling a crown or a solar corona.
Virus began life as a Latin word with the same spelling that meant “poison,” specifically the venom from a snake or spider.
Virus also signified “filthy, slimy,” referring to the foul, filthy and slimy places that caused people to become sick from contact with contaminated water and refuse.
Disease descends from Latin through Old French and originally meant “without ease.” The sense of sickness is not recorded until the very late 14th century.
Another word we’re seeing a hearing a lot these days is quarantine. The first meaning of quarantine, from the Italian quarantina, was a period of 40 days during which a widow had the right to continue living in her deceased husband’s house that was to be seized for debt.
Soon the word took on a related meaning — the 40 days in which a ship suspected of harboring disease had to remain in isolation. The arbitrary number was based on the notion that after 40 days, the disease on board would either have run its course and ended any chance of contagion or would have burst forth its ghastly fury. Finally, quarantine broadened to signify any period of sequestering, and the reference to “40” has vanished.
Then there’s the word vaccinate. For centuries, smallpox was a scourge of humanity, scarring and killing millions. Edward Jenner, a British doctor, noticed that milkmaids did not generally get smallpox and theorized that the pus in the blisters that these women developed from cowpox protected them from the more virulent smallpox. In 1796, Jenner found that inoculating people with a serum containing the lymph gland fluid of cows infected with cowpox virus prevented the similar smallpox. That’s why vaccine, vaccination and vaccinate contain the Latin name for “cow,” vacca.
***
One more thing, the corona virus is one of many corona viruses. Humans will have been carrying one or many in their lives throughout each individuals history of coughs colds and flu. That so much is known about it lends itself to be experimented on throughout its known history, and therefore very much in attention. That science has been able to replicate it for study and add
'go faster' stripes to it, while 'souping up' its engine is undeniable. What has been done with the souped version, and why, is the question. Michaela.
Edited by Paul Ahlquist, University of Wisconsin–Madison, Madison, WI, and approved September 19, 2018 (received for review July 6, 2018)
Significance
Coronaviruses (CoVs) are important pathogens for humans and domestic animals. The development of effective countermeasures against CoVs requires an understanding of the host pathways that regulate viral gene expression and the viral subversion mechanisms. However, little is known about how the stability of viral mRNAs is controlled. We show that the nonsense-mediated decay (NMD) pathway, which primarily targets aberrant cellular mRNAs for degradation, also induced the degradation of CoV mRNAs that are of cytoplasmic origin. Our study further suggests the importance of CoV-induced inhibition of the NMD pathway, mediated by a viral protein, for efficient CoV replication. The present study highlights an interplay between the NMD pathway and CoVs that modulates viral replication by controlling the stability of viral mRNAs.
Abstract
Coronaviruses (CoVs), including severe acute respiratory syndrome CoV and Middle East respiratory syndrome CoV, are enveloped RNA viruses that carry a large positive-sense single-stranded RNA genome and cause a variety of diseases in humans and domestic animals. Very little is known about the host pathways that regulate the stability of CoV mRNAs, which carry some unusual features. Nonsense-mediated decay (NMD) is a eukaryotic RNA surveillance pathway that detects mRNAs harboring aberrant features and targets them for degradation. Although CoV mRNAs are of cytoplasmic origin, the presence of several NMD-inducing features (including multiple ORFs with internal termination codons that create a long 3′ untranslated region) in CoV mRNAs led us to explore the interplay between the NMD pathway and CoVs. Our study using murine hepatitis virus as a model CoV showed that CoV mRNAs are recognized by the NMD pathway as a substrate, resulting in their degradation. Furthermore, CoV replication induced the inhibition of the NMD pathway, and N protein (a viral structural protein) had an NMD inhibitory function that protected viral mRNAs from rapid decay. Our data further suggest that the NMD pathway interferes with optimal viral replication by degrading viral mRNAs early in infection, before sufficient accumulation of N protein. Our study presents clear evidence for the biological importance of the NMD pathway in controlling the stability of mRNAs and the efficiency of replication of a cytoplasmic RNA virus.
Coronaviruses (CoVs) cause a variety of diseases in humans and domestic animals. Most human CoVs usually cause mild to moderate respiratory infections, with the exception of severe acute respiratory syndrome CoV (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV) that cause serious respiratory illness in humans and represent a major public health threat with the potential to inflict massive economic losses (1⇓⇓⇓–5). Currently, there are no approved vaccines and therapeutic agents against human CoVs. Studies that lead to a comprehensive understanding of CoV gene expression strategies and host interactions will provide the necessary knowledge required for the development of new and effective measures to control CoV replication.
CoVs belong to the order Nidovirales, in the family Coronaviridae, and are currently classified into four genera: alpha, beta, gamma, and delta CoVs. CoV is an enveloped virus that carries a large (∼30-kb) positive-sense RNA genome, which is structurally polycistronic, containing multiple open reading frames (ORFs) (6) (SI Appendix, Fig. S1). CoV particles carry a helical nucleocapsid, which is a complex of the viral genomic RNA and the nucleocapsid protein, N, enclosed in an envelope composed of the viral envelope proteins, S, M, and E. After infection, the genomic RNA is released into the cytoplasm and is translated to produce two large polyproteins encoded in gene 1, which occupies the 5′ two-thirds of the genome with two partially overlapping ORFs (SI Appendix, Fig. S1). The two polyproteins are processed by viral proteases to generate 15 or 16 nonstructural proteins, most of which are required for viral RNA synthesis (7). In addition to mRNA 1 (the intracellular form of genomic RNA), several subgenomic mRNAs are synthesized in infected cells (SI Appendix, Fig. S1). These subgenomic mRNAs encode viral structural proteins and accessory proteins, the latter of which are not essential for virus replication in cell culture but play a role in viral pathogenicity (8, 9). CoV mRNAs have a common 3′ end, constituting a 3′-coterminal nested-set structure; the 5′ end of all CoV mRNAs carry a common ∼70-nt leader sequence (SI Appendix, Fig. S1) (6). Accordingly, most of the CoV mRNAs, except for the smallest subgenomic mRNA, have multiple ORFs. Because only the 5′-most ORF in viral mRNAs is, in principle, used for translation, most of the CoV mRNAs have a long 3′ untranslated region (UTR); in the case of genomic RNA, the length of the 3′ UTR is ∼10 kb (SI Appendix, Fig. S1). Although steady progress has been made in understanding CoV gene expression strategies, there are still considerable gaps in our knowledge about the CoV–host interactions involved in the regulation of viral mRNA stability and viral gene expression.
No comments:
Post a Comment
Note: only a member of this blog may post a comment.